As with other emerging viruses, the manner in which Ebolavirus outbreaks appear seemingly from nowhere merely adds to their terror. What is especially intriguing about the current outbreak in West Africa is that it's the wrong virus in the wrong place: as a strain of Zaire Ebolavirus, we would expect to see this virus in central Africa. The modern way to answer the 'where did it come from' question is to compare the genomic sequences of different viruses, and this was one of the first things done for the Guinea isolates.
The problem is where to root the tree. There have been two schools of thought on why outbreaks of Ebola in Africa happen where they do. Firstly, it's simply a case that Ebola is widely distributed and it is a unfortunate event, such as butchering wild animals, that results in spillover from animals into humans. An alternative, is that Ebola is spreading in a wave across Africa, resulting in outbreaks as it progresses, breaking through at points of weakness. For the latter hypothesis we would expect all of the viruses to relate in a straightforward and logical manner.
 |
| Crest of a wave: a figure depicting the hypothesised spread of Ebolavirus in a wave-like fashion. Source |
Drawing a straightforward tree with the sequences from the current outbreak along with the available genome sequences of all Ebolaviruses confirms that the virus causing the outbreak in western Africa is a divergent strain of Ebola Zaire, suggesting it arose in central Africa. However, it's out on its own relative to the main Ebola Zaire clade.
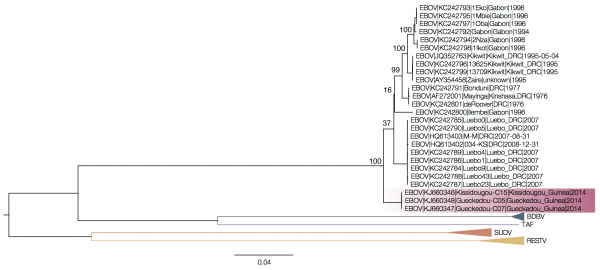 |
| The Ebolavirus tree: assembling a tree using all species of Ebolavirus results in the Guinea sequences separated from the remainder of the Ebola Zaire clade. Source |
The key, it seems, appears to be to remove the intergenic sequences that separate the coding sequences in the virus. When you do this, and concatenate the coding sequences, the sequences from the Guinea outbreak sit in the middle of the Ebola Zaire clade. If you do the same with the intergenic sequences alone you get a tree with similar topology. The issue the authors encounter is that there is no good place from which to root the tree - the other Ebolavirus species are essentially too distant. Combined with the fact that the viruses are always evolving, this all makes it difficult to see what's really happening.
One alternative is to use time, in combination with the estimated rate of evolution, as a way in which to organise the sequences. Using this molecular clock approach results in an intriguing figure whereby the Guinea sequences do indeed come out up top, where we would expect them to be.
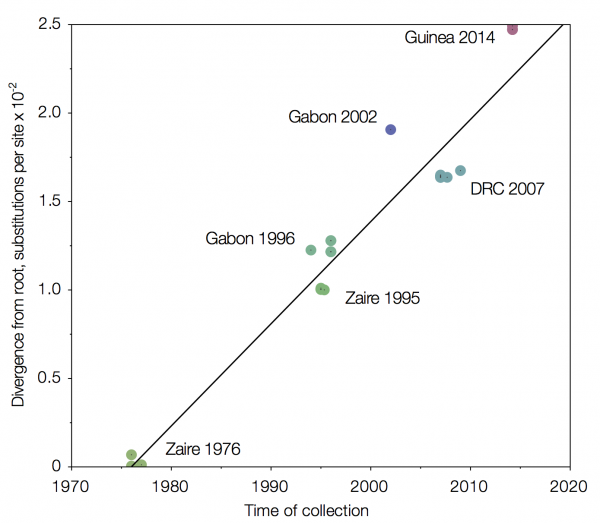 |
| Using a molecular clock to arrange Ebola Zaire sequences places Guinea 2014 furthest from the first isolation in 1976. Source |
One thing that this approach also allows is an estimation as to when the current virus diverged from the central African sequences. Dudas and Rambaut estimate 2002, whereas a separate analysis by Calvignac-Spencer et al, also using molecular clocks to root the Ebola Zaire clade, suggest either 1999 or 2001 (depending upon the assumptions made).
The current outbreak is unprecedented in scale, and it's hard to believe that, when the outbreak is over, there will not be a large study using many sequences. This in turn should give some more clues about how the Ebola wave crashed upon Western Africa.